#012 Principal Component Analysis (PCA)

Highlight: In this post, we will discuss the Principle Component Analysis (PCA). PCA is the bedrock dimensionality reduction technique for probability and statistics. It is commonly used in Data Science and Machine Learning Applications to deal with high-dimensional data that have some statistical distribution. Our goal is to use PCA and uncover the low-dimensional patterns to build models. Before we learn about PCA, we need to understand several steps. First, we will learn about the modes of projections, then we learn about the variance and covariance matrix, then we will learn about eigenvectors and eigenvalues, and finally, we will learn about PCA as a whole. In the following sections, we will discuss these modules in detail through lots of examples and cool illustrations.
Tutorial Overview:
- What is dimensionality reduction?
- What are variance and covariance matrices?
- PCA and a transformation matrix
- Math behind PCA
- Implementation of PCA in Python
1. What is dimensionality reduction?
PCA is also known as a dimensionality reduction method. That means that our data is represented in a higher dimension and we want to see the same information in lower dimensions.
Consider a simple problem. Suppose we have a bunch of friends standing and we want to take a picture of them. Now the question is which angle is best to take a picture? This means that we are interested in finding the best angle to capture a picture so that all of the faces are clearly shown in the picture. Intuitively, we will not take their picture from the back or side view. The angle which will encompass all their faces will be one from the front, and that will be the optimal angle to take the picture. Remember this example because it explains what the variance of the data is. In this case, we have an example where the 3D model is projected into a 2D image. In order to preserve the necessary information, we need to find a way how to map this 3D data into a 2D.

That is what the essence of dimensionality reduction is. We are somehow taking a picture of our data and in this process, we try to keep as much information as possible. For example, let’s consider the data in an image given below:

Here we have a dozen red circles. Each circle is represented in a 2D plane with two coordinates \((x, y) \). Then a picture of the data could be for example projecting it over one line. Then, we can see two different projections. Intuitively which of the two projections seems better? When we compare them, it seems that the second one is better because the points are more spaced out and all points are clearly visible. The goal of dimensionality reduction is to obtain the optimal line to protect the data.

You may wonder: why does this matter? To answer this, consider the following example. We have a data set with multiple columns with the following information that describes the vehicle.

There are several columns here. We want to reduce the number of columns and keep the compressed information. If we say that the first three features are related to the size of a vehicle then maybe we can put them together into something called a size feature. On the other hand, the second two are related to speed so we can put them together into something called a speed feature. This is how dimensionality reduction can be illustrated. We are reducing the number of columns from five to two, but we are still capturing useful information.
We can visualize this if we look at another example. We have a graph in the figure below where we have the number of seats on the horizontal axis and the size of a vehicle on the vertical axis. We are trying to connect those into one feature. We can see that our statement was correct i.e. the bigger the vehicle the more seats it has.

Now we can add a new component called “size feature” that can be recorded along the blue line. Then, we can project the vehicle perpendicular to the line. After doing this our dataset changes. It is much simpler because now it changes from two-dimensional to one-dimensional. Now, every car only has one number attached to it which is a “size feature”.

2. What are variance and covariance matrices?
Consider that we have three weights that are the same, and we want to balance them. How do we find the perfect point of balance for these three weights? We can imagine a wall on the left, measure the distance between every point and the wall, and calculate the average of these points as you can see in the example below:

The next question that we need to answer is, how to differentiate between these sets of points which we can see in the next figure? They have the same mean so we cannot use this method. However, if we take a close look at the figure below, the red points are closer to each other than the green points. So, we need to find out what is the measure of deviation from the mean value. For that, we calculate the variance. As the first step, we can measure the distance from each point to the center. The second step will be to calculate the variance. We will add those numbers and calculate their average. Note that when we take a distance that goes towards the left, we are subtracting its coordinates and we are going to get a negative number. So, to avoid that we need to square these numbers and get a positive number because the distance is always positive. We can see that the variance is the measure of how our data set is spread out.

We can see that the second variance is much bigger than the first one. The next question is: What is the variance for the 2D data set? We can calculate this by finding how much is spread in the horizontal direction and how much is spread in the vertical direction. So basically, we send all points to the horizontal axis and then calculate the variance of these points (x-variance), and then we do the same by sending all points to the vertical axis (y-variance). In this way, we have two numbers, x-variance, and y- variance, and they both measure how much the set is spread out.

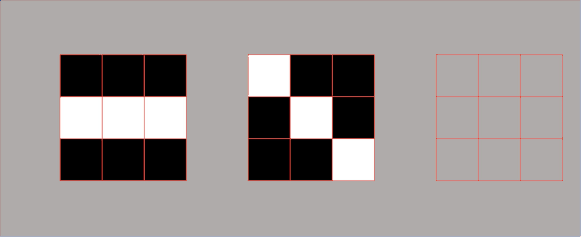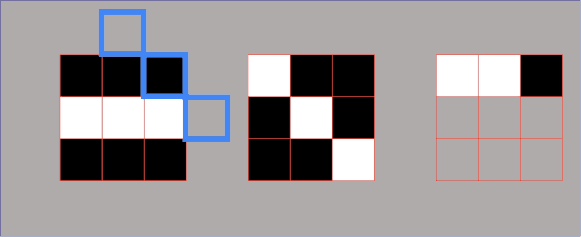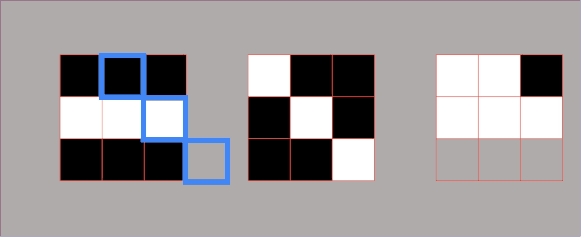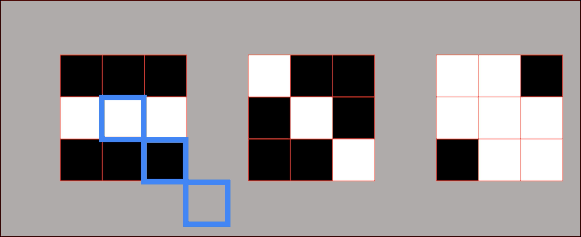
However, there are some cases where we need to introduce a third metric. In the next example, we can see two sets of points that are fundamentally very different. But if we calculate their variances, we can see that they are the same for both sets. So, how can we describe that these two sets are different? For this, we will use a metric known as covariance. It is a metric that will assist us to differentiate between these two sets. And one number that can do that is the product of coordinates. We can see in the following example that product of coordinates is positive for point \(A \) and \(C \) and negative for points \(A_{1} \) and \(B_{1} \). So, the covariance is the sum of the products of coordinates divided by the average.

In the following image, we can see that points on the left are negatively correlated, whereas points on the right are positively correlated. Now, we will calculate the covariance of the following data set.

By looking at the points above you can see that they all cancel out. So, the sum of all these coordinate multiplications is 0. This makes sense because this data set is not positively or negatively correlated. Therefore, by looking at the type of the given set we can assume its covariance. Consider the image below in which you can see that the first set looks like an inverted diagonal, so it got negative covariance. The second one looks as it is well centered, so its covariance is probably very close to zero. The third one has a positive covariance because it looks like a forward diagonal. It is important to remember that covariance is used in PCA.

3. PCA and a transformation matrix
Now we are ready to start a discussion about PCA. We will learn how to find the perfect projection for a given set of points. The first step when working with PCA is to center the data. This means that we will take the average of the coordinates along the \(x \) and the \(y \) direction, find that center of mass and move that center to the origin of the coordinate system. Next, we will create a \(2\times 2 \) covariance matrix called \(C \). In the top left corner, we will put the \(x \) variance (the variance on the horizontal axis) and on the bottom right corner we will put the \(y \) variance (the variance on the vertical axis). In the other two off-diagonal element spots we will write the covariance. For our example data set we can say that the spread along the \(x \) axes is longer than the spread along the \(y \) axes. So the variance may be equal to 9 in the \(x \) direction and to 3 in the \(y \) direction. Also, we will say that the covariance is equal to 4. We know that it is a positive number because it looks like a forward diagonal.

Now, we can explore how this defined covariance matrix \(C \) can transform our data. First, we can see \(C \) as a linear transform matrix. In the following example, we will explain what this linear transformation does, and for more information check out our post – Linear transformations and matrices.
In the image below we can see a unit circle on the left with five points defined by coordinates. When we apply linear transformation each of these points will be mapped in the new point that is calculated by simple multiplication. Now, when we calculate the transformation of all points on the coordinate system on the left and send them to the coordinate system on the right, we can see a pattern. If we look at the unit circle, we can see that it will be transformed into an ellipse. It almost seems like we are stretching the plane in a diagonal direction. That is really what a linear transformation does.

If we take a closer look at the image above we can say that by applying linear transformation with matrix \(\Sigma \), almost all vectors in the plane of the circle will be somehow modified (scaled or rotated). However, we do know that there are two special vectors called eigenvectors that are changed by a scalar factor when that linear transformation is applied to it. For these two vectors, \(i \) and \(j \) applying linear transformation is the same as multiplying that vector with a scalar. In other words, they will be scaled by some value. In our example eigenvector \(i \) will be scaled by 11 and eigenvector \(j \) will be scaled by 1 (it will remain the same).
We can write this formula in the following way. You can see that \(i \) is the eigenvector and \(\lambda \) is the eigenvalue.


Now, we are ready to explain PCA. First, we start with a data set and we zero-center it. Second, we calculate the covariance matrix. The covariance matrix has eigenvectors and eigenvalues which will give direction and magnitude. It is important to highlight that the eigenvectors and eigenvalues preserve most of the information about our data set. Every eigenvector has a corresponding eigenvalue. Eigenvectors will provide information about in what direction the data set is spread and eigenvalue will tell us how much variance there is in that data set. Also, note that when we have a symmetric matrix the eigenvectors \(i \) and \(j \) vectors are perpendicular.

Now, if we look at the image above we can see that eigenvector with magnitude 11 is more dominant. That means that if we would have to preserve information about our data set we pick information along eigenvector \(i \) where the variance is more scattered (the axis that carries the most amount of information). So, we can consider this entire line of the eigenvector \(i \) and project every point of our data set on it. In that way, we turned our two-dimensional data set into a one-dimensional data set. In this process, we obtained a new variable called the first principal component that represents the linear combination of our data set points.

On the other hand, if we project all points of the data set along the \(j \) eigenvector we will obtain the second principal component. However, since the goal of PCA is to reduce the dimensionality of a data set while preserving as much information as possible, the second principal component can be ignored.
4. Math behind PCA
We can represent PCA as a statistical interpretation of the SVD. The principal component analysis provides a data-driven hierarchical coordinate system. So, it gives a hierarchical coordinate system based on data to represent the statistical variations in our data sets.
So, we will start with a data matrix \(X \). It has a bunch of measurements from independent experiments and here we will represent those experiments as big row factors \(x_{1} \), \(x_{2} \), and so on. Here, each row vector \(x \) is essentially measurement from a single experiment. This individual measurement might be the demographic information from a specific human: age, height, weight, sex, or race. Then, \(x_{1} \) will represent person 1, \(x_{2} \) will represent person 2 and so on.

We assume that this data \(x \) has some statistical distribution. It is not deterministic because there is some statistical variability to this information. Our goal is to uncover the dominant kind of combinations of features that describe as much of the data as possible. So, because this is the statistical interpretation of the SVD there are a few steps that we need to cover.
1. Calculating the mean row
Step one in this procedure is to compute the mean row (the average row).
$$ \bar{x}=\frac{1}{n} \sum_{j=1}^{n} x_{j} $$
2. Calculating the average matrix
In the next step, we will find the average matrix. The average matrix \(\bar{X} \) is calculated by taking a vector of ones and multiplying it by the mean row vector \(\bar{x} \). So, we create \(n \) copies of \(\bar{x} \) and that’s our \(\bar{X} \) average matrix.

3. Subtracting the average matrix from the data
In the second step, we will subtract the average matrix from our data.
$$ B= X-{\bar{X}} $$
We call this mean-centered data. We will be modeling this data matrix \(X \) assuming that it is a zero-mean Gaussian.
4. Calculating the covariance matrix of mean-centered data matrix
Next, we will compute the covariance matrix of this mean-centered data matrix. The covariance matrix of \(B \) is:
$$ C=B^{T}B $$
5. Computing the eigenvectors and eigenvalues of the covariance matrix
Next, we will compute the eigenvectors and eigenvalues of this covariance matrix \(C \). That is related both to the singular vectors of \(X \) and also to its principal components. So, let us see how we can compute the eigenvalues and eigenvectors. We can write the following formula:
$$ V_{1}^{T} B^{T} B V_{1} $$
The largest eigenvector of this matrix \(B^{T} B \) is \(V_{1} \). After we compute \(V_{1} \) we will compute \(V_{2} \), \(V_{3} \) and so on. In this way, we will get a matrix as given below:

Consider the next equation where we show what principal components are.

So, we can calculate matrix \(T \) by multiplying our mean subtracted data \(B \) with eigenvector \(V \). Then, we can say that the matrix \(T \) consists of principal components, and these vectors \(V \) of these eigenvectors are called the loadings.
In this way, we decomposed this matrix into directions of maximal variance (just like in the singular value decomposition) called the principal components and the loadings where the loadings represent how much of each of those principal components each of the experiments has.
$$ B = U \Sigma V^{T} $$
If we took the singular value decomposition of the mean subtracted data:
$$ B = U \Sigma V^{T} $$
Then \(T \) would be equal to \(B \) times \(V \). And we can see that \(B \) times \(V \) is simply \(U \) times \(\Sigma \). Therefore, this equation is also a representation of the principal components.
$$ T = U \Sigma $$
The important thing about is the eigenvalues or the singular values in \(\Sigma \) is that they give you an indication of the amount of the variance of this data set that these principal components capture. We can say that if we only want to describe this high dimensional data in terms of the first two principal components and the first two vectors of loadings, we would be able to compute how much of the variance is captured in those first two eigenvalues of this \(D \) matrix.
We can say that these eigenvalues lambda are equal to the square of the singular values and it is equal to the variance of that principal component in the data. And if want to know how much variance is being captured in the first several components, we would write the following equation:
$$ \lambda =\sigma ^{2} $$

So, we can see what is the fraction of variance captured by our first lambdas divided by the total of all of the variants in the data set. For example, we might decide to keep only as many principal components as are needed to explain 95% of the variance. So that would give us criteria for how many principal components we need to keep.
Now let’s see how we can implement the PCA in the Python code.
5. Implementation of PCA in Python
For our first experiment in Python we are goin to calculate principal components of a 10000 data points which are spreaded in a 2D space. First, we will import the necessary libraries. Then, we will define the center of the data, principal axes and we are going to rotate data by angle theta. After we compute the average of the data we will subtract that center with all the data. This process brings the data back to the origin. After that, we compute the SVD, which tells us the rotation (\(U \) matrix), and how stretched is the datam(singular values \(S \)). Then we are going to figure out what is the 1st and 2nd standard confidence interval, based on the information from the SVD. When we take the SVD of the matrix of data we find the \(S \) matrix (sigma). It has two elements that tell us that the first principal direction has a lot of variances and the second has a small variance. On the other hand, the \(U \) matrix tells us how this distribution is rotated at what angle are those principal components.
import matplotlib.pyplot as plt
import numpy as np
x_center = np.array([-1, 1]) # Center of data (mean)
x_center = np.diag(x_center)
sig = np.array([0.5, 2]) # Principal axes
sig = np.diag(sig)
theta = np.pi/3 # Rotate by pi/3
R = np.array([[np.cos(theta), -np.sin(theta)], # Rotation matrix
[np.sin(theta), np.cos(theta)]])
nPoints = 10000
# Rotate, Stretch and smash, Center data
X = np.matmul(np.matmul(R, sig), np.random.randn(2,nPoints)) + np.matmul(x_center, np.ones((2,nPoints))) # Create 10,000 points
fig = plt.figure(figsize=(16, 8))
ax1 = fig.add_subplot(121)
ax1.plot(X[0,:],X[1,:], '.', Color='k')
ax1.grid()
plt.xlim((-8, 7))
plt.ylim((-4, 7))
Xavg = np.mean(X,axis=1) # Compute mean (average, center of data)
B = X - np.tile(Xavg,(nPoints,1)).T # Mean-subtracted data
# Find principal components (SVD)
U, S, VT = np.linalg.svd(B/np.sqrt(nPoints)) # Rotation U, Stretch S
S = np.diag(S)
plt.plot(X[0,:],X[1,:], '.', Color='k') # Plot data to overlay PCA
plt.grid()
plt.xlim((-8, 7))
plt.ylim((-4, 7))
theta = 2 * np.pi * np.arange(0,1,0.01)
# 1-std confidence interval
Xstd = np.matmul(np.matmul(U, S), np.array([np.cos(theta),np.sin(theta)])) # U * S * unit circle and it maps into the distribution
plt.plot(Xavg[0] + Xstd[0,:], Xavg[1] + Xstd[1,:],'-',color='r',LineWidth=3)
plt.plot(Xavg[0] + 2*Xstd[0,:], Xavg[1] + 2*Xstd[1,:],'-',color='r',LineWidth=3)
plt.plot(Xavg[0] + 3*Xstd[0,:], Xavg[1] + 3*Xstd[1,:],'-',color='r',LineWidth=3)
# Plot principal components U[:,0]S[0] and U[:,1]S[1]
plt.plot(np.array([Xavg[0], Xavg[0]+U[0,0]*S[0,0]]),
np.array([Xavg[1], Xavg[1]+U[1,0]*S[0,0]]),'-',color='cyan',LineWidth=4)
plt.plot(np.array([Xavg[0], Xavg[0]+U[0,1]*S[1,1]]),
np.array([Xavg[1], Xavg[1]+U[1,1]*S[1,1]]),'-',color='cyan',LineWidth=4)
plt.show()
As you can see in the graph above he red circles are the standard confidence intervals, and the blue lines are principal components.
Ovarian cancer dataset
The next goal is to apply PCA to the Ovarian cancer dataset. It is a comprehensive dataset that contains nearly all the PLCO study data available for ovarian cancer screening, incidence, and mortality analyses. The dataset contains 216 patients and 4000 markers measured for every patient. Broken in groups, the first 108 have cancer and the last 108 don’t have cancer. Because 4000 points are far too many to plot and understand, the PCA is going to learn some genetic sequences. When we SVD this (\(216\times4000 \)) matrix we get a (216, 216) matrix, diagonal S, and VT matrix. The first row of VT is a gene sequence that captures the most variation between every patient, the second captures the second most, and so on. Now instead of visualizing all 4000 gene sequences, we can plot the first few sequences, lower dimension.
To download the Ovarian cancer dataset we will run the following commands:
- !wget https://www.dropbox.com/s/bal044f83ezxiia/ovariancancer_obs.csv
- !wget https://www.dropbox.com/s/r52sn5gmkd3y797/ovariancancer_grp.csvNow, we can load the dataset, compute the SVD and plot the results.
!wget https://www.dropbox.com/s/bal044f83ezxiia/ovariancancer_obs.csv
!wget https://www.dropbox.com/s/r52sn5gmkd3y797/ovariancancer_grp.csv
import matplotlib.pyplot as plt
import numpy as np
from mpl_toolkits.mplot3d import Axes3D
# Load dataset
features = np.loadtxt('/content/ovariancancer_obs.csv', delimiter=',')
# Load labels (Cancer, Normal)
f = open('/content/ovariancancer_grp.csv', "r")
labels = f.read().split("\n")
# Compute Singular Value Decomposition (SVD)
U, S, VT = np.linalg.svd(features, full_matrices=0)
fig1 = plt.figure(figsize=(16, 8))
ax1 = fig1.add_subplot(121)
ax1.semilogy(S,'-o',color='c') # We can see that
ax2 = fig1.add_subplot(122)
ax2.plot(np.cumsum(S)/np.sum(S),'-o',color='r')
plt.show()
We can see from the first plot that a huge part of the variance comes from the first 3 singular values, and as it goes down we have a huge roll-off where there is little information. By keeping the first 10 singular values we can capture a lot of variance between all of the patients. The right plot is the cumulative sum, showing how much variance is captured by the first R singular vectors.
fig2 = plt.figure()
ax = fig2.add_subplot(111, projection='3d')
# We take our first features (observation) matrix and project it
# in to the first 3 principal components.
# Take every patients genetic sequence and dot product it with
# the first three VT rows and get three numbers as the output
# We plot these 3 numbers
for j in range(features.shape[0]):
x = np.matmul(VT[0, :], features[j, :].T)
y = np.matmuk(VT[1, :], features[j, :].T)
z = np.matmul(VT[2, :], features[j, :].T)
if labels[j] == 'Cancer':
ax.scatter(x, y, z, marker='x', color='r', s=50)
else:
ax.scatter(x, y, z, marker='o', color='b', s=50)
ax.view_init(25, 20)
plt.show()
The X’s represent patients with cancer and O’s represent patients without. We can see that with the first 3 principal components, the data is pretty much separated. If we would take the first 5-10 components it would be even better spread out. This way we got a low dimensional, human interpretable way of understanding this information.
PCA on Olivetti faces dataset
The PCA is often applied in face recognition algorithms. That is why we will show how you can apply PCA to images. For this experiment, we will use the Olivetti faces dataset. For that, we can use a built-in python function for PCA, from the sklearn library. We are going to set the n_components argument to 150, which is the number of features.
from sklearn.datasets import fetch_olivetti_faces
images = fetch_olivetti_faces()
fig = plt.figure(figsize=(8, 6))
for i in range(4):
ax = fig.add_subplot(3, 5, i+1, xticks=[], yticks=[])
ax.imshow(images.images[i], cmap=plt.cm.gray)
data = images.data
# Scratch
U0, S0, VT0 = np.linalg.svd(data, full_matrices=0)
# Built-in function
from sklearn.decomposition import PCA
pca = PCA(n_components=150, whiten=True)
pca.fit(data)fig3 = plt.figure()
ax3 = fig3.add_subplot(111)
ax3.semilogy(S0,'-o',color='k')
fig4 = plt.figure(figsize=(12, 8))
ax4 = fig4.add_subplot(111, projection='3d')
for i in range(len(data)):
x = np.matmul(VT0[0, :], data[i])
y = np.matmul(VT0[1, :], data[i])
z = np.matmul(VT0[2, :], data[i])
ax4.scatter(x, y, z, marker='x', c='r', s=50)
plt.show()
We can reconstruct some of the images as given below:
fig = plt.figure(figsize=(16, 6))
for i in range(5):
ax = fig.add_subplot(3, 10, i + 1, xticks=[], yticks=[])
ax.imshow(pca.components_[i].reshape(images.images[0].shape),
cmap=plt.cm.gray)
Summary
In this post, we covered a lot of theory and code. We explained why PCA is still a very important method in data science and machine learning applications when we deal with a large amount of data. We explained what dimensionality reduction means, what is variance and covariance matrix, and we explained in detail the math behind the PCA method. In the next post, we will talk about Eigenfaces.